R for reproducible scientific analysis
Dataframe manipulation with tidyr
Learning Objectives
- To be understand the concepts of ‘long’ and ‘wide’ data formats and be able to convert between them with
tidyr
Researchers often want to manipulate their data from the ‘wide’ to the ‘long’ format, or vice-versa. The ‘long’ format is where:
- each column is a variable
- each row is an observation
In the ‘long’ format, you usually have 1 column for the observed variable and the other columns are ID variables.
For the ‘wide’ format each row is often a site/subject/patient and you have multiple observation variables containing the same type of data. These can be either repeated observations over time, or observation of multiple variables (or a mix of both). You may find data input may be simpler or some other applications may prefer the ‘wide’ format. However, many of R‘s functions have been designed assuming you have ’long’ format data. This tutorial will help you efficiently transform your data regardless of original format.
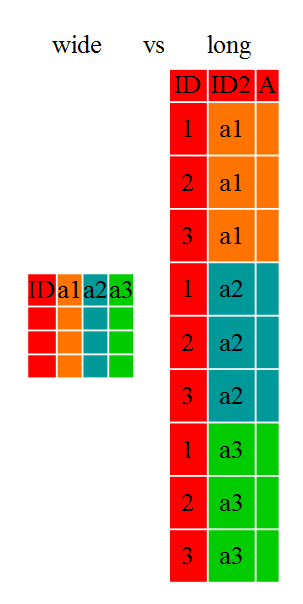
These data formats mainly affect readability. For humans, the wide format is often more intuitive since we can often see more of the data on the screen due to it’s shape. However, the long format is more machine readable and is closer to the formating of databases. The ID variables in our dataframes are similar to the fields in a database and observed variables are like the database values.
Getting started
First install the packages if you haven’t already done so (you probably installed dplyr in the previous lesson):
#install.packages("tidyr")
#install.packages("dplyr")Load the packages
library("tidyr")
library("dplyr")First, lets look at the structure of our original healthData dataframe:
str(healthData)'data.frame': 2255 obs. of 15 variables:
$ id : int 3 4 7 8 10 12 15 17 18 20 ...
$ conscientiousness : num 5.83 7.73 6.5 5.88 4.25 ...
$ extraversion : chr "3.986" "7.016" "2.697" "2.504" ...
$ intellect : num 6.04 6.82 5.53 4.23 4.75 ...
$ agreeableness : chr "4.613" "6.649" "3.087" "4.613" ...
$ neuroticism : chr "3.649" "6.299" "4.091" "3.649" ...
$ sex : chr "Male" "Male" "Male" "Male" ...
$ selfRatedHealth : int 4 5 3 3 4 4 4 4 5 4 ...
$ mentalAdjustment : int 2 3 3 2 2 2 3 1 3 3 ...
$ illnessReversed : int 3 5 4 4 3 5 2 4 5 4 ...
$ health : num 6.74 11.96 8.05 6.48 6.74 ...
$ alcoholUseInYoungAdulthood: int 2 3 2 1 2 2 1 1 1 2 ...
$ education : int 9 8 6 8 9 4 6 7 9 9 ...
$ birthYear : int 1909 1905 1910 1905 1910 1911 1903 1908 1909 1911 ...
$ HIGroup : chr "Group 1" "Group 1" "Group 1" "Group 1" ...
Challenge 1
Is healthData a purely long, purely wide, or some intermediate format?
Sometimes, we have multiple types of observed data. It is somewhere in between the purely ‘long’ and ‘wide’ data formats. We have 2 “ID variables” (id,HIGroup) and 13 “Observation variables”. I usually prefer my data in this intermediate format in most cases despite not having ALL observations in 1 column given that all observation variables have different units. There are few operations that would need us to stretch out this dataframe any longer (i.e. 3 ID variables and 1 Observation variable).
While using many of the functions in R, which are often vector based, you usually do not want to do mathematical operations on values with different units. For example, using the purely long format, a single mean for all of the values of intellect, conscientiousness and health would not be meaningful since it would return the mean of values with 3 incompatible units. The solution is that we first manipulate the data either by grouping (see the lesson on dplyr), or we change the structure of the dataframe. Note: Some plotting functions in R actually work better in the wide format data.
From intermediate to long format with gather()
Until now, we’ve been using the nicely formatted original healthData dataset, but ‘real’ data (i.e. our own research data) may not be so well organized. To demonstrate, let’s engineer a less usefully structure dataset using tidyr’s gather() function.
str(healthData)'data.frame': 2255 obs. of 15 variables:
$ id : int 3 4 7 8 10 12 15 17 18 20 ...
$ conscientiousness : num 5.83 7.73 6.5 5.88 4.25 ...
$ extraversion : chr "3.986" "7.016" "2.697" "2.504" ...
$ intellect : num 6.04 6.82 5.53 4.23 4.75 ...
$ agreeableness : chr "4.613" "6.649" "3.087" "4.613" ...
$ neuroticism : chr "3.649" "6.299" "4.091" "3.649" ...
$ sex : chr "Male" "Male" "Male" "Male" ...
$ selfRatedHealth : int 4 5 3 3 4 4 4 4 5 4 ...
$ mentalAdjustment : int 2 3 3 2 2 2 3 1 3 3 ...
$ illnessReversed : int 3 5 4 4 3 5 2 4 5 4 ...
$ health : num 6.74 11.96 8.05 6.48 6.74 ...
$ alcoholUseInYoungAdulthood: int 2 3 2 1 2 2 1 1 1 2 ...
$ education : int 9 8 6 8 9 4 6 7 9 9 ...
$ birthYear : int 1909 1905 1910 1905 1910 1911 1903 1908 1909 1911 ...
$ HIGroup : chr "Group 1" "Group 1" "Group 1" "Group 1" ...
The tidyr function gather() can ‘gather’ your observation variables into a single variable.
healthData_long <- healthData %>% gather(obsType,obsValues,-id,-HIGroup)
# OR
# healthData_long <- healthData %>% gather(obsType,obsValues,conscientiousness,extraversion,intellect,
# agreeableness,neuroticism,sex,selfRatedHealth,mentalAdjustment,illnessReversed,
# health,alcoholIseInYoungAdulthood,education,birthYear)
str(healthData_long)'data.frame': 29315 obs. of 4 variables:
$ id : int 3 4 7 8 10 12 15 17 18 20 ...
$ HIGroup : chr "Group 1" "Group 1" "Group 1" "Group 1" ...
$ obsType : chr "conscientiousness" "conscientiousness" "conscientiousness" "conscientiousness" ...
$ obsValues: chr "5.825" "7.732" "6.498" "5.881" ...
Here we have used piping syntax which is similar to what we were doing in the previous lesson with dplyr. In fact, these are compatible and you can use a mix of tidyr and dplyr functions by piping them together.
Inside gather() we first name the new column for the new ID variable (obsType), the name for the new amalgamated observation variable (obsValue), then the names of the old observation variable. We could have typed out all the observation variables, but gather also allows the alternative syntax of using the - symbol to identify which variables are not to be gathered (i.e. ID variables).
That may seem trivial with this particular dataframe, but sometimes you have 1 ID variable and 40 Observation variables with irregular variables names. The flexibility is a huge time saver!
Now obsType actually contains the observation type (conscientiousness,intellect, health etc), and obsValue contains the values for that observation for that particular id. Due to the coersion rules we introduced earlier, since some of the observation variables where character data types, all the observations are now represented as strings. As a result of the structure change, we now have many rows per id, where before we had only one row per id. The resulting data.frame is much longer.
From long to intermediate format with spread()
Now just to double-check our work, let’s use the opposite of gather() to spread our observation variables back out with the aptly named spread(). We can then spread our healthData_long to the original intermediate format or the widest format. Let’s start with the intermediate format.
healthData_normal <- healthData_long %>% spread(obsType,obsValues)
dim(healthData_normal)[1] 2255 15
dim(healthData)[1] 2255 15
names(healthData_normal) [1] "id" "HIGroup"
[3] "agreeableness" "alcoholUseInYoungAdulthood"
[5] "birthYear" "conscientiousness"
[7] "education" "extraversion"
[9] "health" "illnessReversed"
[11] "intellect" "mentalAdjustment"
[13] "neuroticism" "selfRatedHealth"
[15] "sex"
names(healthData) [1] "id" "conscientiousness"
[3] "extraversion" "intellect"
[5] "agreeableness" "neuroticism"
[7] "sex" "selfRatedHealth"
[9] "mentalAdjustment" "illnessReversed"
[11] "health" "alcoholUseInYoungAdulthood"
[13] "education" "birthYear"
[15] "HIGroup"
Now we’ve got an intermediate dataframe healthData_normal with the same dimensions as the original healthData, but the order of the variables is different. Let’s fix that before checking if they are all.equal().
healthData_normal <- healthData_normal[,names(healthData)]
all.equal(healthData_normal,healthData) [1] "Component \"id\": Mean relative difference: 0.5459826"
[2] "Component \"conscientiousness\": Modes: character, numeric"
[3] "Component \"conscientiousness\": target is character, current is numeric"
[4] "Component \"extraversion\": 2199 string mismatches"
[5] "Component \"intellect\": Modes: character, numeric"
[6] "Component \"intellect\": target is character, current is numeric"
[7] "Component \"agreeableness\": 2159 string mismatches"
[8] "Component \"neuroticism\": 2153 string mismatches"
[9] "Component \"sex\": 1088 string mismatches"
[10] "Component \"selfRatedHealth\": Modes: character, numeric"
[11] "Component \"selfRatedHealth\": target is character, current is numeric"
[12] "Component \"mentalAdjustment\": Modes: character, numeric"
[13] "Component \"mentalAdjustment\": target is character, current is numeric"
[14] "Component \"illnessReversed\": Modes: character, numeric"
[15] "Component \"illnessReversed\": target is character, current is numeric"
[16] "Component \"health\": Modes: character, numeric"
[17] "Component \"health\": target is character, current is numeric"
[18] "Component \"alcoholUseInYoungAdulthood\": Modes: character, numeric"
[19] "Component \"alcoholUseInYoungAdulthood\": target is character, current is numeric"
[20] "Component \"education\": Modes: character, numeric"
[21] "Component \"education\": target is character, current is numeric"
[22] "Component \"birthYear\": Modes: character, numeric"
[23] "Component \"birthYear\": target is character, current is numeric"
[24] "Component \"HIGroup\": 942 string mismatches"
head(healthData_normal) id conscientiousness extraversion intellect agreeableness neuroticism
1 1 4.815 3.342 3.587 3.087 4.091
2 2 5.32 3.342 5.204 2.069 .
3 3 5.825 3.986 6.044 4.613 3.649
4 4 7.732 7.016 6.821 6.649 6.299
5 7 6.498 2.697 5.527 3.087 4.091
6 8 5.881 2.504 4.234 4.613 3.649
sex selfRatedHealth mentalAdjustment illnessReversed health
1 Female 4 3 3 8.31
2 Female 4 1 3 5.17
3 Male 4 2 3 6.74
4 Male 5 3 5 11.96
5 Male 3 3 4 8.05
6 Male 3 2 4 6.48
alcoholUseInYoungAdulthood education birthYear HIGroup
1 1 5 1913 Group 1
2 1 8 1911 Group 1
3 2 9 1909 Group 1
4 3 8 1905 Group 1
5 2 6 1910 Group 1
6 1 8 1905 Group 1
head(healthData) id conscientiousness extraversion intellect agreeableness neuroticism
1 3 5.825 3.986 6.044 4.613 3.649
2 4 7.732 7.016 6.821 6.649 6.299
3 7 6.498 2.697 5.527 3.087 4.091
4 8 5.881 2.504 4.234 4.613 3.649
5 10 4.254 5.147 4.751 3.850 3.208
6 12 7.508 3.535 6.821 4.613 5.415
sex selfRatedHealth mentalAdjustment illnessReversed health
1 Male 4 2 3 6.74
2 Male 5 3 5 11.96
3 Male 3 3 4 8.05
4 Male 3 2 4 6.48
5 Male 4 2 3 6.74
6 Male 4 2 5 9.01
alcoholUseInYoungAdulthood education birthYear HIGroup
1 2 9 1909 Group 1
2 3 8 1905 Group 1
3 2 6 1910 Group 1
4 1 8 1905 Group 1
5 2 9 1910 Group 1
6 2 4 1911 Group 1
We’re almost there, but the data doesn’t match quite. The output of the head() function shows that each data.frame is sorted differently. To ensure consistent sorting, we can use the arrange() function from the dplyr package.
healthData_normal <- healthData_normal %>% arrange(id)
healthData <- healthData %>% arrange(id)
str(healthData_normal)'data.frame': 2255 obs. of 15 variables:
$ id : int 1 2 3 4 7 8 10 12 14 15 ...
$ conscientiousness : chr "4.815" "5.32" "5.825" "7.732" ...
$ extraversion : chr "3.342" "3.342" "3.986" "7.016" ...
$ intellect : chr "3.587" "5.204" "6.044" "6.821" ...
$ agreeableness : chr "3.087" "2.069" "4.613" "6.649" ...
$ neuroticism : chr "4.091" "." "3.649" "6.299" ...
$ sex : chr "Female" "Female" "Male" "Male" ...
$ selfRatedHealth : chr "4" "4" "4" "5" ...
$ mentalAdjustment : chr "3" "1" "2" "3" ...
$ illnessReversed : chr "3" "3" "3" "5" ...
$ health : chr "8.31" "5.17" "6.74" "11.96" ...
$ alcoholUseInYoungAdulthood: chr "1" "1" "2" "3" ...
$ education : chr "5" "8" "9" "8" ...
$ birthYear : chr "1913" "1911" "1909" "1905" ...
$ HIGroup : chr "Group 1" "Group 1" "Group 1" "Group 1" ...
str(healthData)'data.frame': 2255 obs. of 15 variables:
$ id : int 1 2 3 4 7 8 10 12 14 15 ...
$ conscientiousness : num 4.82 5.32 5.83 7.73 6.5 ...
$ extraversion : chr "3.342" "3.342" "3.986" "7.016" ...
$ intellect : num 3.59 5.2 6.04 6.82 5.53 ...
$ agreeableness : chr "3.087" "2.069" "4.613" "6.649" ...
$ neuroticism : chr "4.091" "." "3.649" "6.299" ...
$ sex : chr "Female" "Female" "Male" "Male" ...
$ selfRatedHealth : int 4 4 4 5 3 3 4 4 4 4 ...
$ mentalAdjustment : int 3 1 2 3 3 2 2 2 3 3 ...
$ illnessReversed : int 3 3 3 5 4 4 3 5 5 2 ...
$ health : num 8.31 5.17 6.74 11.96 8.05 ...
$ alcoholUseInYoungAdulthood: int 1 1 2 3 2 1 2 2 1 1 ...
$ education : int 5 8 9 8 6 8 9 4 6 6 ...
$ birthYear : int 1913 1911 1909 1905 1910 1905 1910 1911 1904 1903 ...
$ HIGroup : chr "Group 1" "Group 1" "Group 1" "Group 1" ...
Now we can see that the data matches, but the datatypes are different for some columns due to the coersion that occured earlier.
healthData_normal$conscientiousness <- as.numeric(healthData_normal$conscientiousness)
healthData_normal$intellect <- as.numeric(healthData_normal$intellect)
healthData_normal$selfRatedHealth <- as.integer(healthData_normal$selfRatedHealth)
healthData_normal$mentalAdjustment <- as.integer(healthData_normal$mentalAdjustment)
healthData_normal$illnessReversed <- as.integer(healthData_normal$illnessReversed)
healthData_normal$health <- as.numeric(healthData_normal$health)
healthData_normal$alcoholUseInYoungAdulthood <- as.integer(healthData_normal$alcoholUseInYoungAdulthood)
healthData_normal$education <- as.integer(healthData_normal$education)
healthData_normal$birthYear <- as.integer(healthData_normal$birthYear)
all.equal(healthData_normal,healthData)[1] TRUE
That’s great! We’ve gone from the longest format back to the intermediate and we didn’t introduce any errors in our code.
Challenge 2
Convert the original healthData data.frame to a wide format which has the 2 original id columns (id and HIGroup), as well as columns for education, birthYear and sex. Combine all other observation columns (conscientiousness,extraversion,intellect etc) into a single pair of columns - one which hold observation type, and one with the observation value.
Solution to Challenge 1
The original gapminder data.frame is in an intermediate format. It is not purely long since it had multiple observation variables (pop,lifeExp,gdpPercap).
Solution to Challenge 2
healthData_longish <- healthData %>% gather(obsType,obsValues,-id,-HIGroup,-education,-birthYear,-sex).png)